
Initially, the outer loop assigns k=1L and executes its statement which is to the inner loop, meanwhile the inner for loop assigns l=1L and therefore prints k=1, l=1.For instance, let’s take the following code: This is more beneficial if we wish to extract a specific value from the corresponding row and column index. Nested for loops are used to manipulate a matrix by making a specific setting to a specific value and considered as a foundation skill in R Programming. Flowchart representing the steps of Nested ‘For’ Loop: when there is no value it returns to end. Flowchart Structureīelow flowchart shows the R for Loop structures: In the below diagram for each value in the sequence, the loop gets executed. We can use numeric as well as character indices.
Nested Loops are primarily meant for multi-dimensional array storage purposes which is widely used by the data scientist. In the above general syntax, we could see two loop statements. Here variable implies iteration value used in a sequence and sequence is a set of values or objects that could be numbers or characters. So I would suggest changing your for loop to construct a list of ames directly.Hadoop, Data Science, Statistics & othersĮxpression // expression statements can be a single statement of R or group of statements. What if there is some object Buffer123 that you were playing around with and don't want merged? Then merge this with your APbuff: merge(APbuff, buffer_all, by="SLL_ID")Īll this being said, I don't recommend relying on object names like this. Now you just need to merge this list of ames, by the variable name they have in common SLL_ID, in the following non-obvious manner: buffer_all <- Reduce(function(x, y) merge(x, y, by="SLL_ID"), mget(ls(pattern = 'Buffer+'))) This gives you the objects themselves in what is called a list, in this case a list of ames. mget to the rescue: mget(ls(pattern = 'Buffer+'))) Gives you only the objects whose names start with "Buffer" and one or more numbers.īut this only gives you the object names.
Gives you all your objects, but this: ls(pattern = 'Buffer+')) The ls function lets you search the environment's objects by pattern.
One way would leave your for loop unchanged and will use the fact the object names all have the string "Buffer" in them followed by a number.
#FOR LOOP IN R WITH DATA FRAME PLUS#
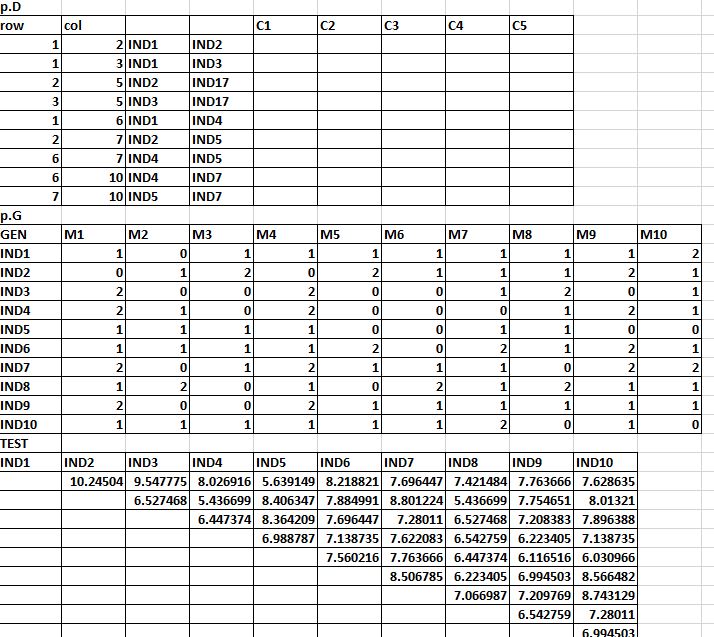
(Appears to be a ame with one plus tens or hundreds of columns.) But if you insist, there are a few ways of doing this. I'm not so sure what you actually want is going to end up being a good idea. V <- setNames(aggregate(v$cumulative, by=list(v$SLL_ID), FUN=max), c("SLL_ID", nam)) V <- setNames(aggregate(APdist$temp,by=list(APdist$SLL_ID, APdist$temp), FUN=sum), c("SLL_ID","buffer","cumulative")) This is what my current for loop looks like: for(i in seq(50,250, by=50)) Buffer50, Buffer100, etc.) with APbuff, so that I have the SLL_ID 1:187, the SLL_Values column and the cumulative sum column (Buffer50, Buffer100, etc.) from each data frame from the for loop (i.e. I want to merge each of the data frame outputs from my for loop (i.e. Example of what the data frame looks like: Data frame name: APbuff I have another data frame called APbuff with 187 observations and two columns (an ID 1:187 and values). Example of what a Buffer# data frame looks like: Data frame name: Buffer50 Each data frame has 187 observations and two columns (an SLL_ID 1:187 and cumulative sum, with a column name that is the same as the data frame). I have a for loop that outputs data frames named Buffer50, Buffer100.,Buffer250.
Hello, I'm very new to R, so please bear with me.


 0 kommentar(er)
0 kommentar(er)
